
Sampling for skin microbiome .
OMNIgene® SKIN is specifically designed for the collection of skin samples for microbiome analysis. The kit is suitable for skin samples from dry, moist and sebaceous areas. The efficient preservation solution efficiently stabilizes microbial DNA, preventing distortion of the microbial profile due to microbial growth or DNA degradation. The easy handling is optimal for self-sampling by patients.
Hard Facts .
- Ideal for self-sampling by patients/study participants
- Suitable for dry, damp or sebaceous skin removal areas
- Perfect preservation of microbial profiles (<30 days).
- Suitable for many downstream applications (16S, ITS2 and WGS)
- Stabilizes the microbial profile against temperature fluctuations (-20°C to +50°C)
Applications .
The Swab-based OMNIgene®-SKIN (OMR-140) kit provides a perfect snapshot of the human skin microbiome. The collection of high-quality microbial DNA is also successful from skin areas of different textures.
Bouevitch, A., Macklaim, J., & Le François, B. (2020). OMNIgene®-SKIN (OMR-140): an optimized collection device for the capture and stabilization of the human skin microbiome. DNA Genotek https://www.dnagenotek.com/ROW/pdf/PD-WP-00067.pdf
OMNIgene SKIN - OMR-140 .
Product overview .
| Attribute | OMNIgene SKIN (OMR-140) |
|---|---|
| Volume of stabilizing solution | 1 ml |
| Optimized for bacteriome and mycobiome profiling | ✔ |
| Freeze/thaw and temperature fluctuation stability | -20ºC to +50ºC |
| Validated for 16S, ITS2 and metagenomic WGS sequencing | ✔ |
| Microbiome profile stability at room temperature | 30 days |
| Suitable for all skin types (dry, wet, sebaceous) | ✔ |
| Validated method incorporating low bioburden components | ✔ |
| Integrated 2D barcode for sample traceability and workflow efficiency | ✔ |

Advantages .
- Self-collection of high-quality DNA from skin sites with low microbial biomass (dry, moist or sebaceous).
- Stabilization of bacterial and fungal profiles at the point of collection from all skin types.
- Sample stability of 30 days at ambient temperature eliminates the need for immediate freezer storage and cold chain during transport
- Realistic representation of microbial composition by minimizing distortions caused by microbial growth and DNA degradation.
- Isolation of high quality DNA suitable for PCR, 16S, ITS2 and metagenomic shotgun sequencing.
- Intuitive handling improves donor compliance and sample collection quality
- Perfect stabilization of the microbial profile against temperature fluctuations during shipment and storage
- Enables downstream analysis even at low population densities
More information .
-
Manufacturer websiteManufacturer website
-
Instruction manual (engl.)Instruction manual (engl.)
-
Data SheetData Sheet
Unbiased microbial profiles .
Even after 30 days of storage at room temperature, there are no significant taxonomic changes in the microbial profile of the sample.
From: Bouevitch, A., Macklaim, J., & Le François, B. (2020). OMNIgene®-SKIN (OMR-140): an optimized collection device for the capture and stabilization of the human skin microbiome. DNA Genotek. https://www.dnagenotek.com/ROW/pdf/PD-WP-00067.pdf
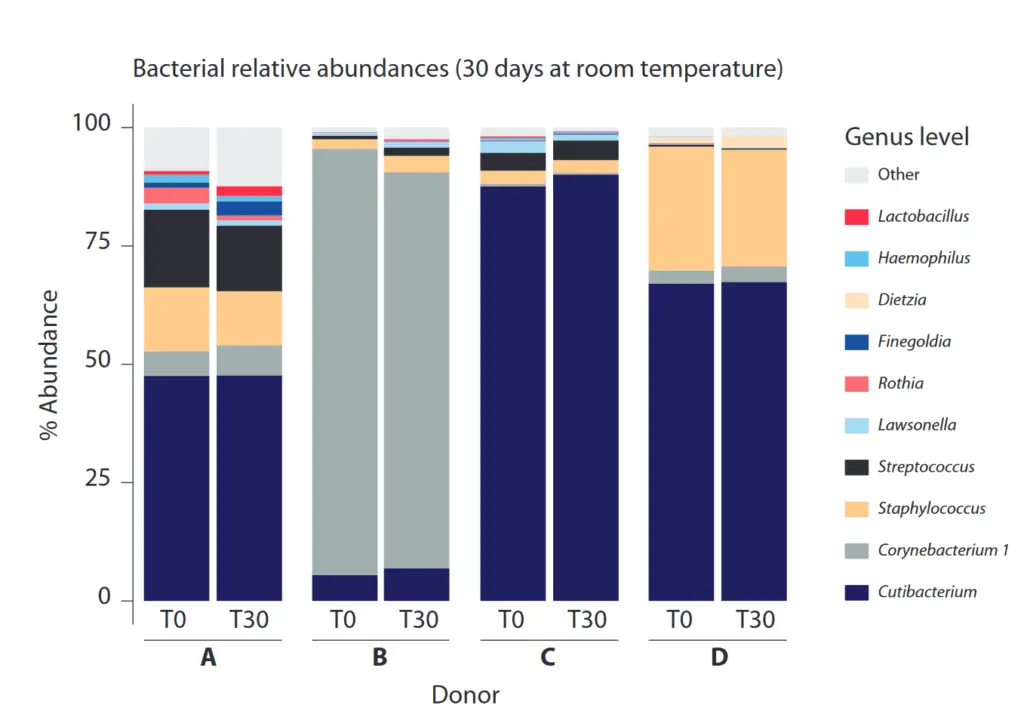
Stability of taxonomic profile in OMNIgene SKIN samples stored at room temperature for 30 days. The 16S rRNA gene from T0- and T30-extracted samples was amplified with primers for V3-V4, and the relative abundance plot was constructed from the filtered amplicon sequence variant (ASV) data classified using the SILVA database and aggregated to genus level or the lowest assignable taxonomic level, as indicated in parentheses.
Taxonomic profiles from different sampling sites. .
16S taxonomic profiling showed that OMNIgene-SKIN captured bacterial taxa known to be associated with specific skin sites (Figure Left). For example, Cutibacterium (formally known as Propionibacterium) was mainly associated with sebaceous skin sites (face and scalp), while Corynebacterium and Staphylococcus were the two most prevalent genera on toe pads (moist skin sites). Also, the taxonomic ITS2 profiles showed that Malassezia was found in high relative sebaceous and dry skin of most donors in high relative abundance majority of donors, while a greater diversity of fungal genera was found a greater diversity of fungal genera on toe webs (Figure Right).
From: Bouevitch, A., Macklaim, J., & Le François, B. (2020). OMNIgene®-SKIN (OMR-140): an optimized collection device for the capture and stabilization of the human skin microbiome. DNA Genotek. https://www.dnagenotek.com/ROW/pdf/PD-WP-00067.pdf

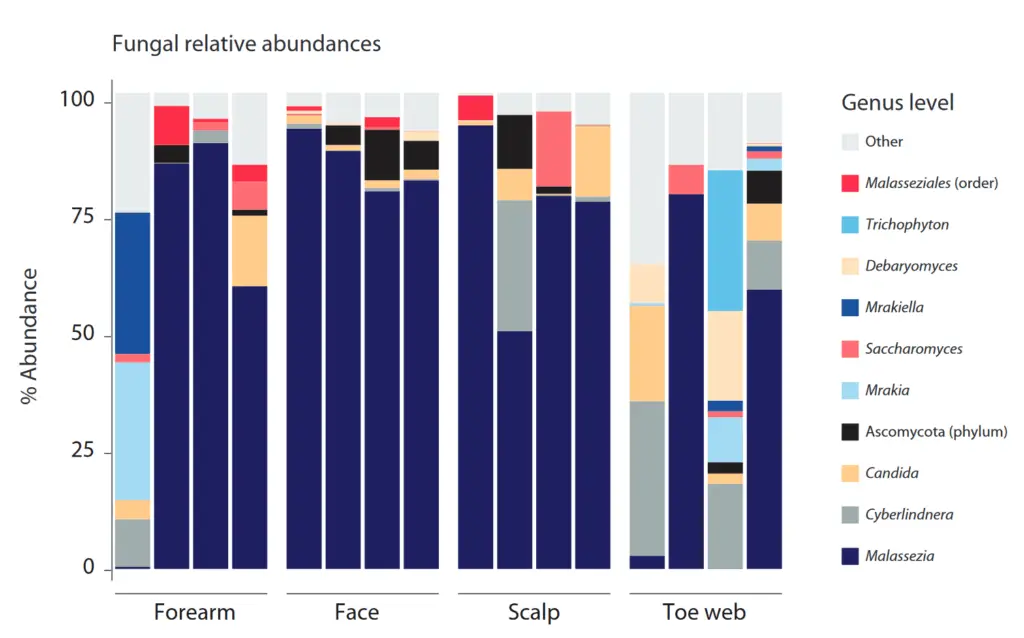
Bacterial and fungal taxonomic profiles of microbial skin samples collected from the forearm (dry), face and scalp (sebaceous), and toenails (wet) using OMNIgene-SKIN.
LEFT: The 16S rRNA gene was amplified with primers for V3-V4. The relative abundance plot was generated from the filtered amplicon sequence variant (ASV) data classified using the SILVA database and at the genus level or the lowest or the lowest assignable taxonomic level, as indicated in parentheses.
RIGHT: For fungal analysis, the ITS2 region was amplified by PCR and relative abundance was generated from filtered ASV data classified using the UNITE database and at the genus level or lowest assignable taxonomic level as indicated in parentheses. Both in (A) as well as in (B), the 10 most abundant taxa from 4 representative donors are shown, while the remaining Reads.
grouped as “other”.
Instructions for sampling by the user .
Step-by-step instructions for self-collection of specimen material.

