cDNA synthesis .
Our primaREVERSE BASIC Reverse Transcription Kit contains everything you need for cDNA synthesis from purified total RNA.
The proprietary reverse transcriptase with reduced RNase-H activity and increased thermostability guarantees high specificity and yield of cDNA synthesis. The included Oligo(dT) and Random Hexamer primers allow you to very easily optimize the protocol according to your needs.
Hard Facts .
- Ready-to-use complete kit incl. Primer and DNase
- Suitable for templates up to 17 kb
- High yield due to reduced RNase-H activity
- Complete reverse transcription in under 10 minutes
- For use with specific primers, random hexamers or oligo(dT) primers.
- For complex transcripts the RT can be raised up to 65°C.

Applications .
Fast and easy cDNA synthesis with only one kit. The primaREVERSE RT kit offers high sensitivity and specificity. In addition, it is very flexible and provides high yields with oligo(dT), random hexamers or gene-specific primers.
Gene expression levels in tissues or different cell types can be determined by detecting specific mRNA. For this purpose, the total RNA is first transcribed into cDNA. In the subsequent qPCR with gene-specific primers the respective expression level is determined.
In RNA virus diagnostics a cDNA library is created prior to sequencing. In addition to pathogen detection itself, it can also be used to generate mutation profiles of pathogens.
As part of an RNA-seq workflow, mRNA enrichment can be performed using reverse transcription in conjunction with oligo(dT) primers. This allows both mRNA selection and RT to be performed in one step, which is particularly useful for small amounts of RNA. It should be noted that a 3′ bias may occur during subsequent sequencing. The result would be an increased number of reads for the 3′ end of the RNAs.
The features .
Simple workflow for maximum cDNA yield .
With the primaREVERSE RT kit, you can transcribe RNA into cDNA in less than 15 minutes.
You do not need to pipette on ice, as the product includes an RNase inhibitor that prevents degradation of total RNA.

Content .

Content of the kit:
| Component | Concentration |
|---|---|
| 10x DNase Incubation Buffer | 10x |
| DNase Enzyme Mix | 5 U/µl |
| Reverse Transcriptase | 200 U/µl |
| 5x Reaction Buffer | 5x |
| dNTP Mix | 10 mM |
| Oligo-(dT)20 Primer | 100 µM |
| Random Hexamer Primer | 100 µM |
| RNase Inhibitor | 40 U/µl |
| RNase-free water | – |
Kit Packaging:
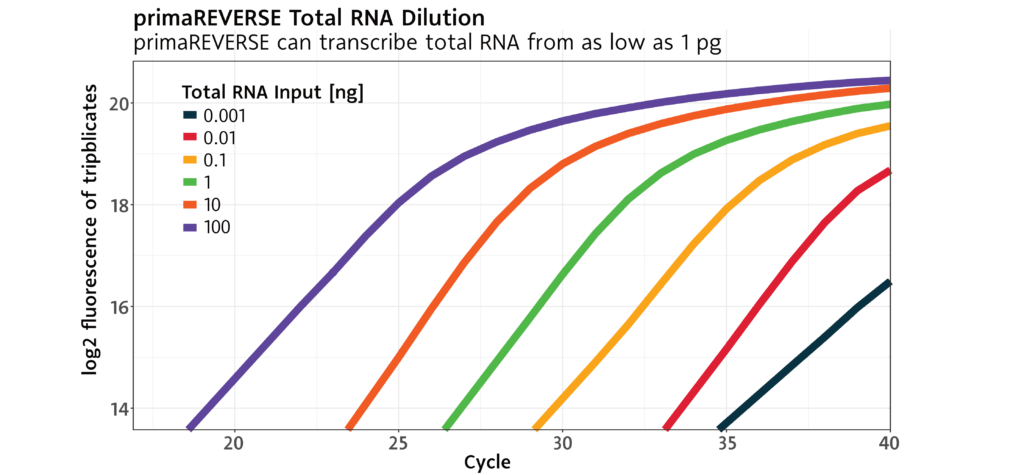
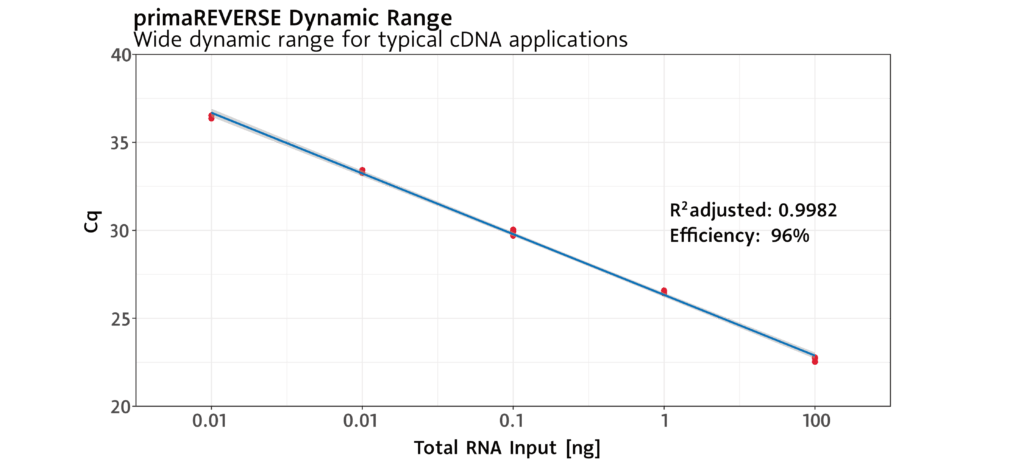
Sensitive to minute amounts of RNA .
The primaREVERSE Reverse Transcription Kit enables cDNA synthesis even from samples with the smallest amounts of RNA.
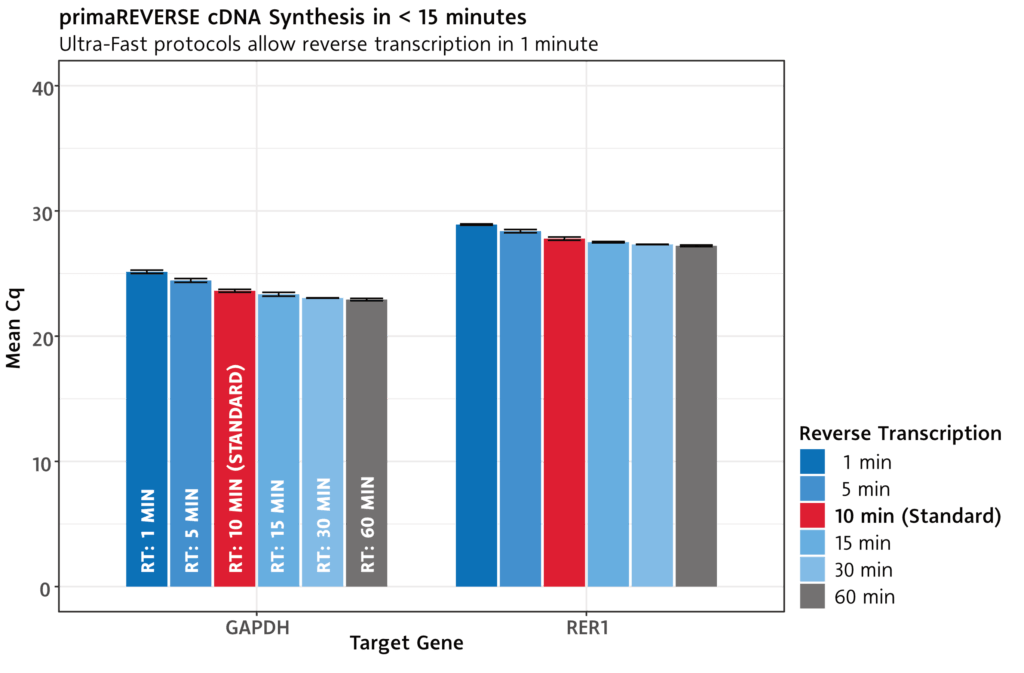
From RNA to cDNA in less than 15 minutes .
The primaREVERSE RT Kit allows transcription from RNA to cDNA in less than 15 minutes – with full performance.
With our proprietary ultra-fast reverse transcriptase, cDNA synthesis can even be shortened to as little as 1 minute, with only a minimal reduction in performance.
For complex transcripts, reverse transcription can be extended up to 60 minutes.
The table with ID 0 not exists.
Primer included .
- Random Hexamers
- OligoDT(20)
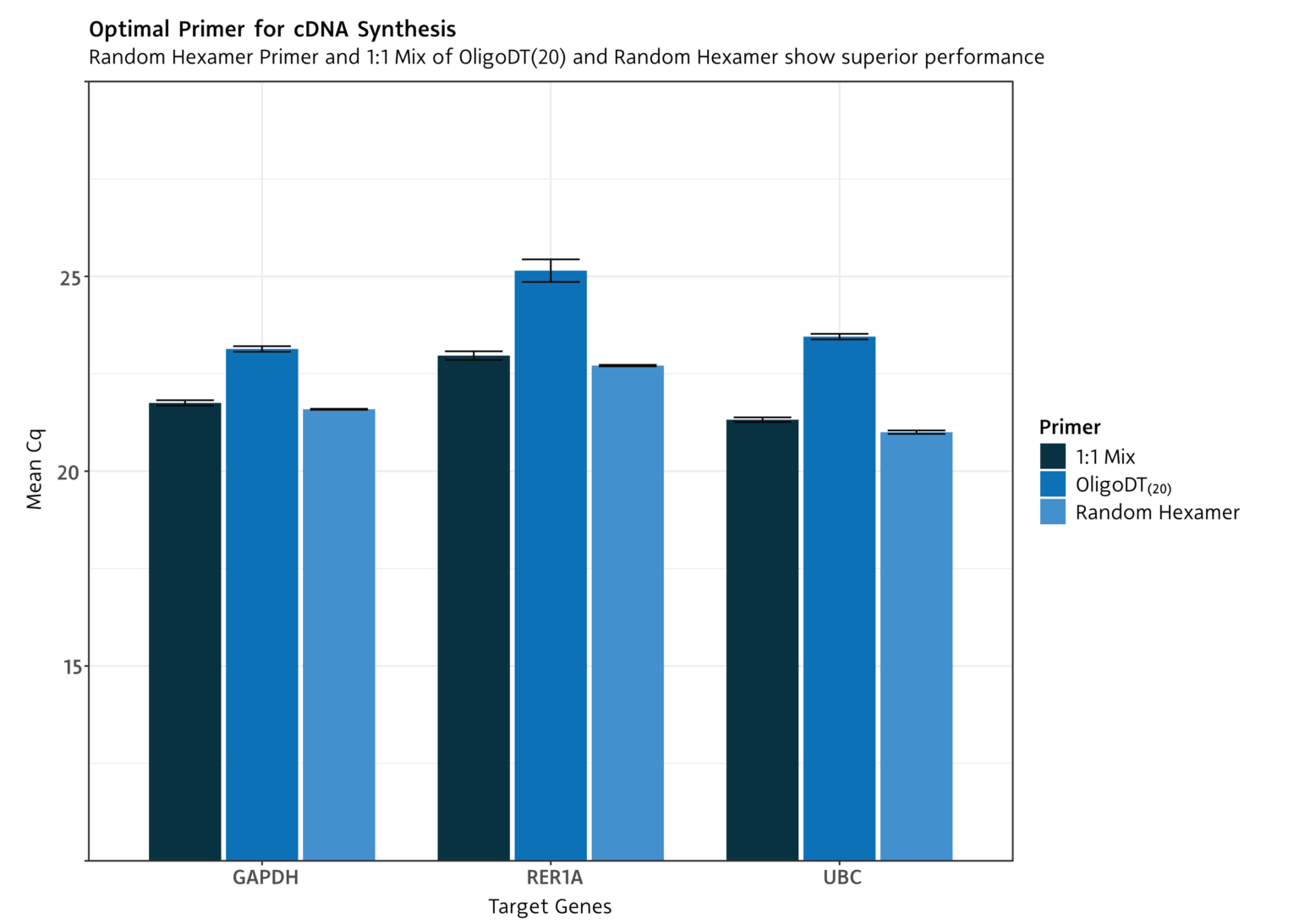
The primaREVERSE RT kit contains Random Hexamer and OligoDT(20) primers so you can make the best choice for your assay.
In many cases, a 1:1 mixture of random hexamer and oligoDT(20) primer or random hexamer primers alone will give you the best results. However, in some assays, it may also be useful to use oligoDT(20) primers that bind to the poly-A tail of the mRNA.
The primaREVERSE RT kit is also ideally suited for the use of gene-specific primers. Details can be found in the manual.
Which primer should I use ?
For most assays, we recommend using either a 1:1 mixture of random hexamer and OligoDT(20) primer or random hexamer primer alone.
gDNA Removal for more accurate target identification .
The primaREVERSE RT kit contains a DNase enzyme mix for removal of genomic DNA.
Removal of genomic DNA from total RNA is generally recommended because even primers that do not bind on genomic DNA can generate a signal in qPCR, see the following figure.

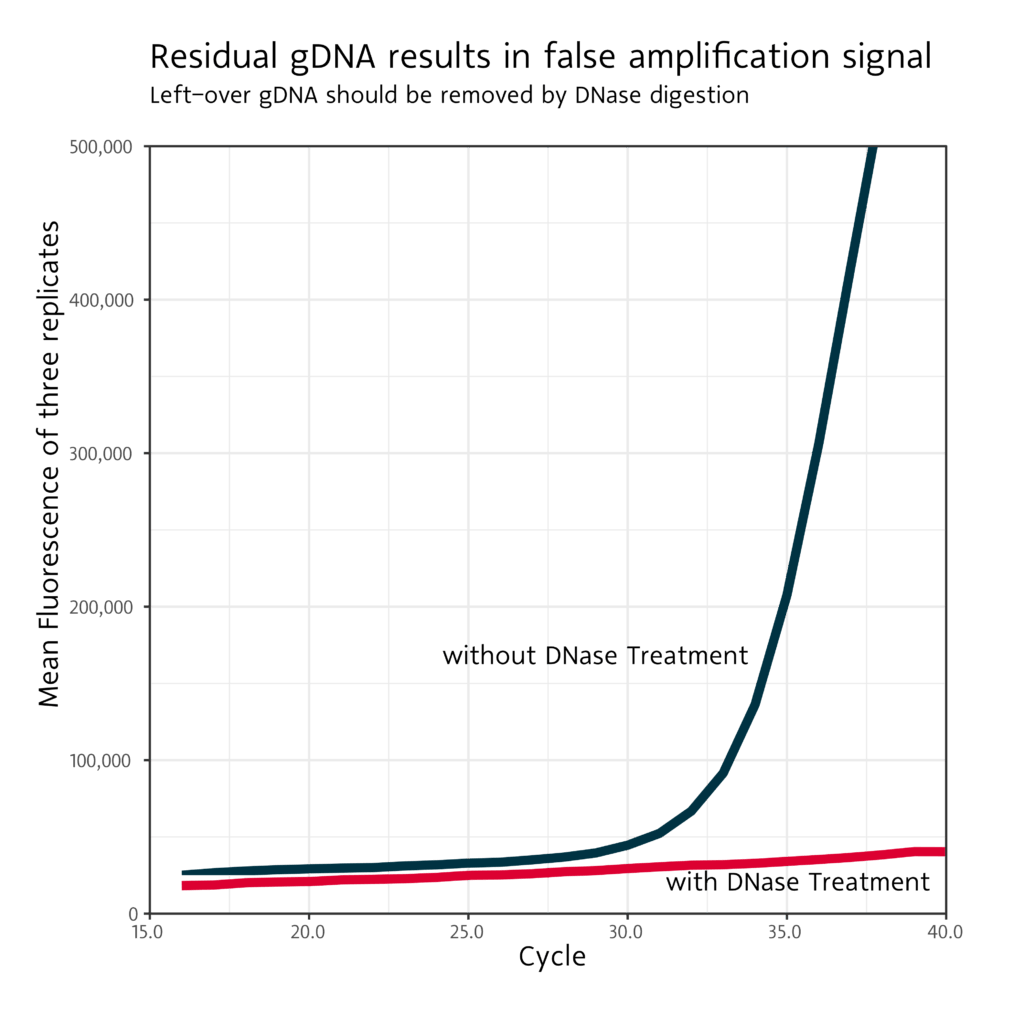
Without DNase digestion, the remaining genomic DNA can also generate a signal.
With a prior DNase digest, non-specific amplification is prevented and the signal is not distorted.
Figure 1:
qPCR curve plot for GAPDH with RNA-specific primers and an input amount of 1000 ng total RNA. Without DNase digestion, the genomic DNA still present in the sample leads to an amplification signal.
DNase digestion can be used to prevent unwanted amplification.
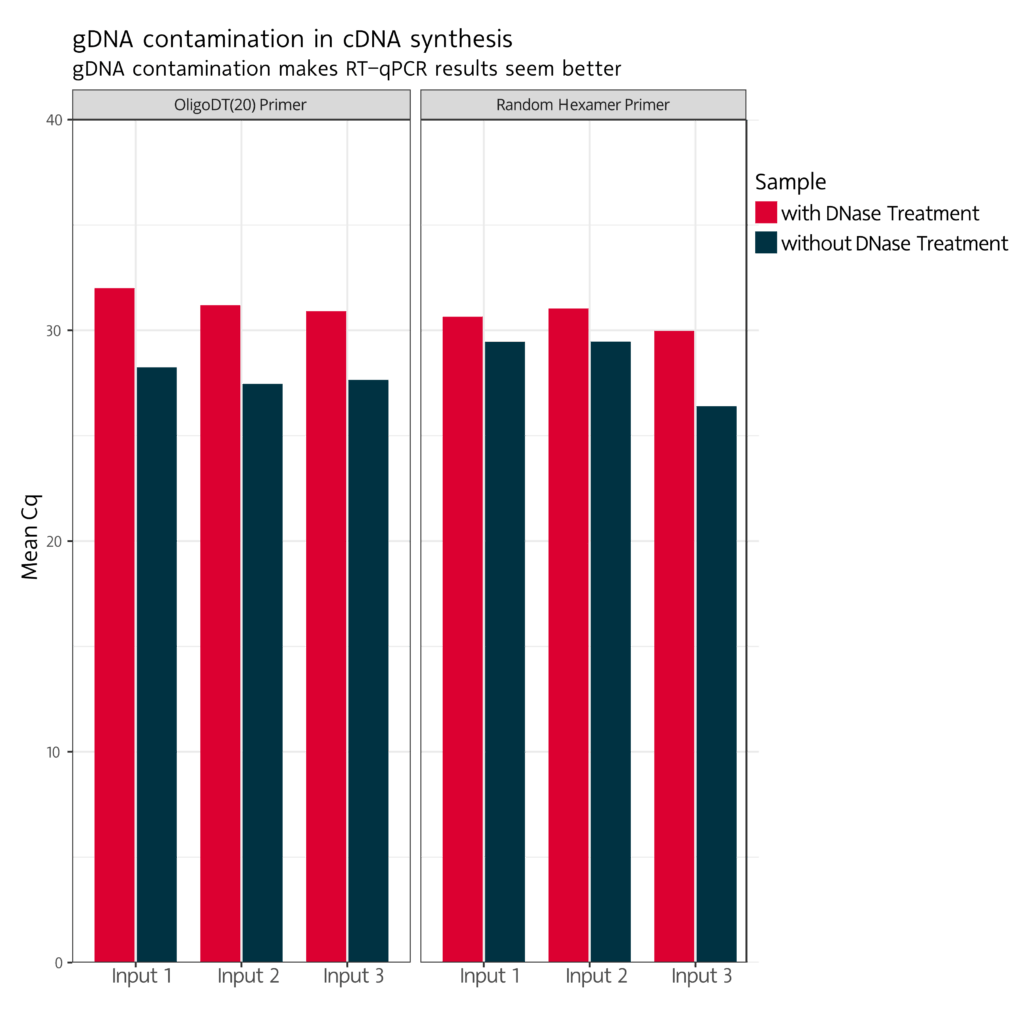
Without DNase digestion, the remaining
genomic DNA may generate a signal,
which belies a higher gene expression.
With a DNase digest
a non-specific amplification is being
prevented and the signal is not distorted.
Figure 2:
qPCR results (Cq values) with RNA-specific primers and an input amount of 10 ng total RNA. Reverse transcription was performed using OligoDT or random hexamer primers.
Without DNase digestion, the qPCR signal will be distorted. Gene expression appears higher than it actually is.
High efficiency of reverse transcriptase even at high temperatures .
The modified M-MuLV transcriptase allows longer RT runs at temperatures up to 65°C – perfect for difficult template RNAs.
Made in Germany .
We develop and produce our enzymes and master mixes exclusively ourselves in Germany. You benefit from this in several ways:
- Very short delivery times
- Fast and competent support
- ISO certified production
- High-end quality control
- Customized fillings or product adaptations

Reverse Transcription Kit .
Downloads .
The table with ID 0 not exists.
Primer/RNA Mix
The table with ID 0 not exists.
Final Reaction Mix
The table with ID 0 not exists.